Multi-'omics Atlas Project
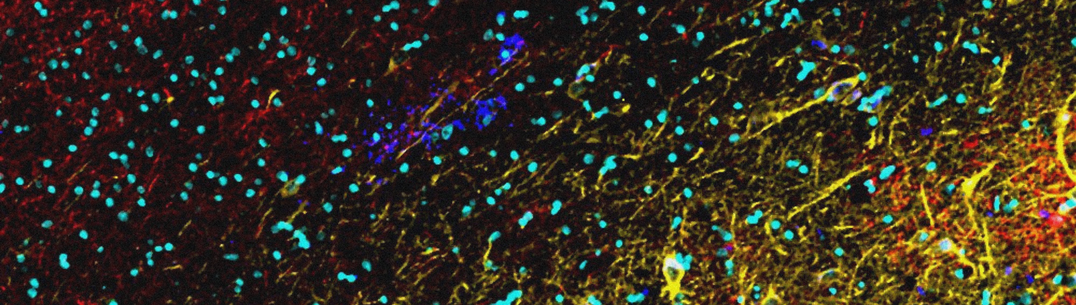
Neurodegenerative diseases arise from interactions between cells throughout the brain; they are not a cell- or regionally- autonomous process. The pathologies evolve from primary dysfunction through secondary pathology or adaptive changes. The Multi-‘omics Atlas Project, a UK DRI Director’s Initiative, has generated a well characterised, clinically annotated set of Alzheimer’s disease and control brain tissue collected using approaches harmonised across contributing brain banks. Multi-‘omics (e.g., genomics, transcriptomics, proteomics and lipidomics) data acquired across different brain regions and across different stages of the disease have been generated and tools for their characterisation are available through an ‘open science’ platform to encourage collaborative science addressing major clinical challenges of dementia.
The UK DRI Multi-‘omics Atlas Project is an open resource dedicated to the comprehensive, multi-omic mapping of the cellular pathology of Alzheimer’s disease over representative stages in its evolution (MAP AD).
This project uses an unprecedented range of advanced techniques to examine tissue from different regions of the Alzheimer’s Disease brain. This will gain a much fuller understanding of key cell characteristics, including what influences the genes of individual cells to be expressed, and the role of proteins and other factors on their function.
2 column block
Key Objectives
To establish a UK DRI Multi-‘omics Atlas Project, an open resource dedicated to the comprehensive, multi-omic mapping of the cellular pathology of Alzheimer’s disease over representative stages in its evolution.- To use well-phenotyped tissue as a foundation with standardised data acquisitions.
- To build open and integrated datasets which are linked to the brain from which they are derived.
- To create a platform which is extensible to include other platforms and other investigators as the project progresses.
Enquiries
Please address enquiries about this program to:
Scientific Project Lead
Dr Johanna Jackson
Johanna.jackson@imperial.ac.uk
View Professor Matthews' professional web page and research publications
View Dr Jackson's professional web page and research publications